4. Workflow and Git
An essential skill for using R effectively and reproducibly is having a clear, standardized workflow. I’ll walk through one using GitHub here, and you should always use a consistent workflow when you work in R.
Git and GitHub are commonly-used tools for version control and code-sharing. In order to get started, you’ll need to create a free account on GitHub: https://github.com/
Next, you’re going to want to verify that git is installed and ready to go on your computer. In order to do that, you should follow this guide: https://happygitwithr.com/install-git.html (although you can use the terminal tab in RStudio instead of the shell to follow the instructions in 6.1 if you like — I find that much easier).
Now, you should be configured to follow the following work flow:
- Create a meaningfully-named GitHub repository
- Create a new project in R and link it to this repo
- Store all related files in this project folder with meaningfully-named, consistent subfolders (such as a “data” folder that stores all of your data)
- Use the Git tab in R to consistently knit, commit, and push
Let’s walk through this:
I. Create a meaningfully-named GitHub repository
In GitHub, click on Repositories and then click New:
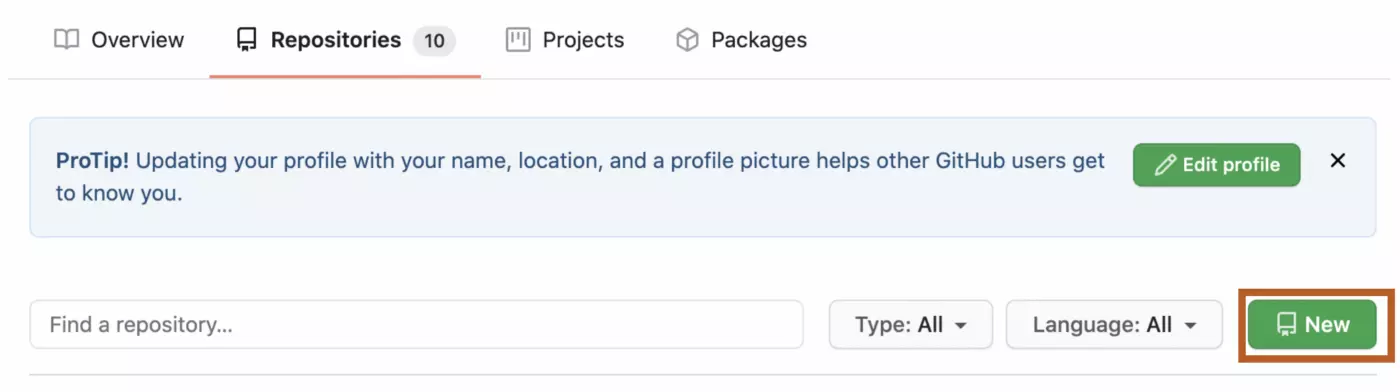
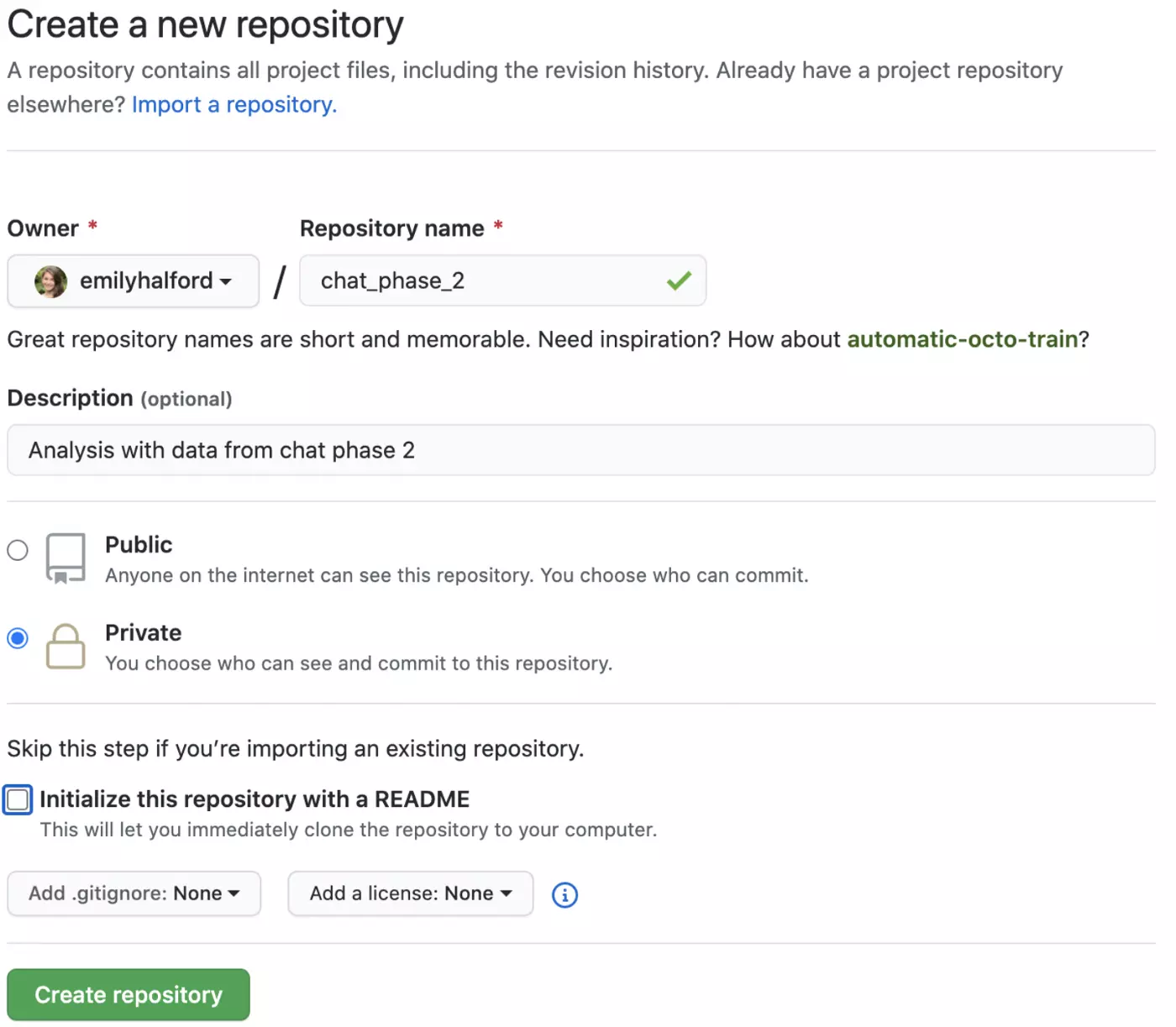
From there, you can give your repo a name and brief description. This repo contained data from the second phase of a study using online chat data, and has therefore been named “chat_phase_2.” I know that this name will allow me to find the repo if I come back to it several months later, and that my collaborators will be able to keep track of it as well.
Your repo can either be public (anyone can find your repo and its contents) or private (only you and designated collaborators can see it and its contents). There are many great uses for public repos (e.g. sharing packages you create, sharing cool projects you create) but if you are using data that contains confidential information and/or are conducting research that has not yet been published, you ALWAYS want to use a private repo. You can change these designations later, but should always be careful about what information you make public.
Typically, you will want to create a README file to guide yourself and anyone else who might access your repo through its contents. A good README outlines the contents of your repo, as well as providing a brief rationale for the work you’ve done.
Click “create repository”, and then copy the link that will appear on the following page:
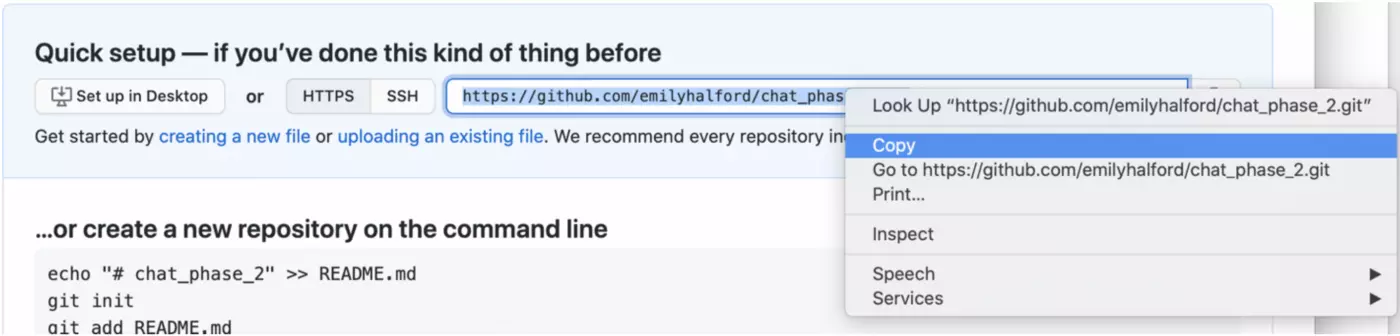
II. Create a new project in R and link it to this repo
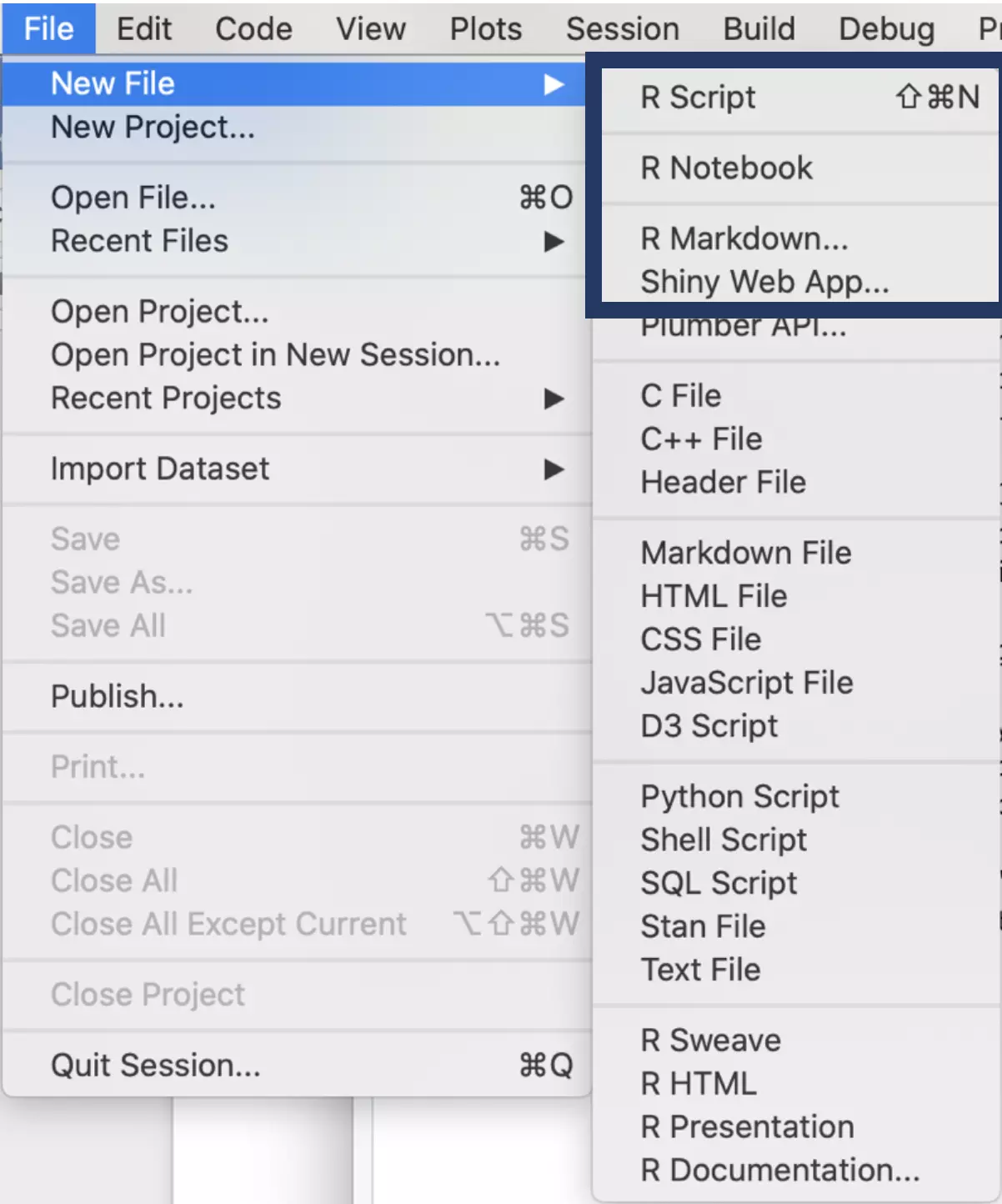
In R, click File → New Project
Next, select “Project with version control”:
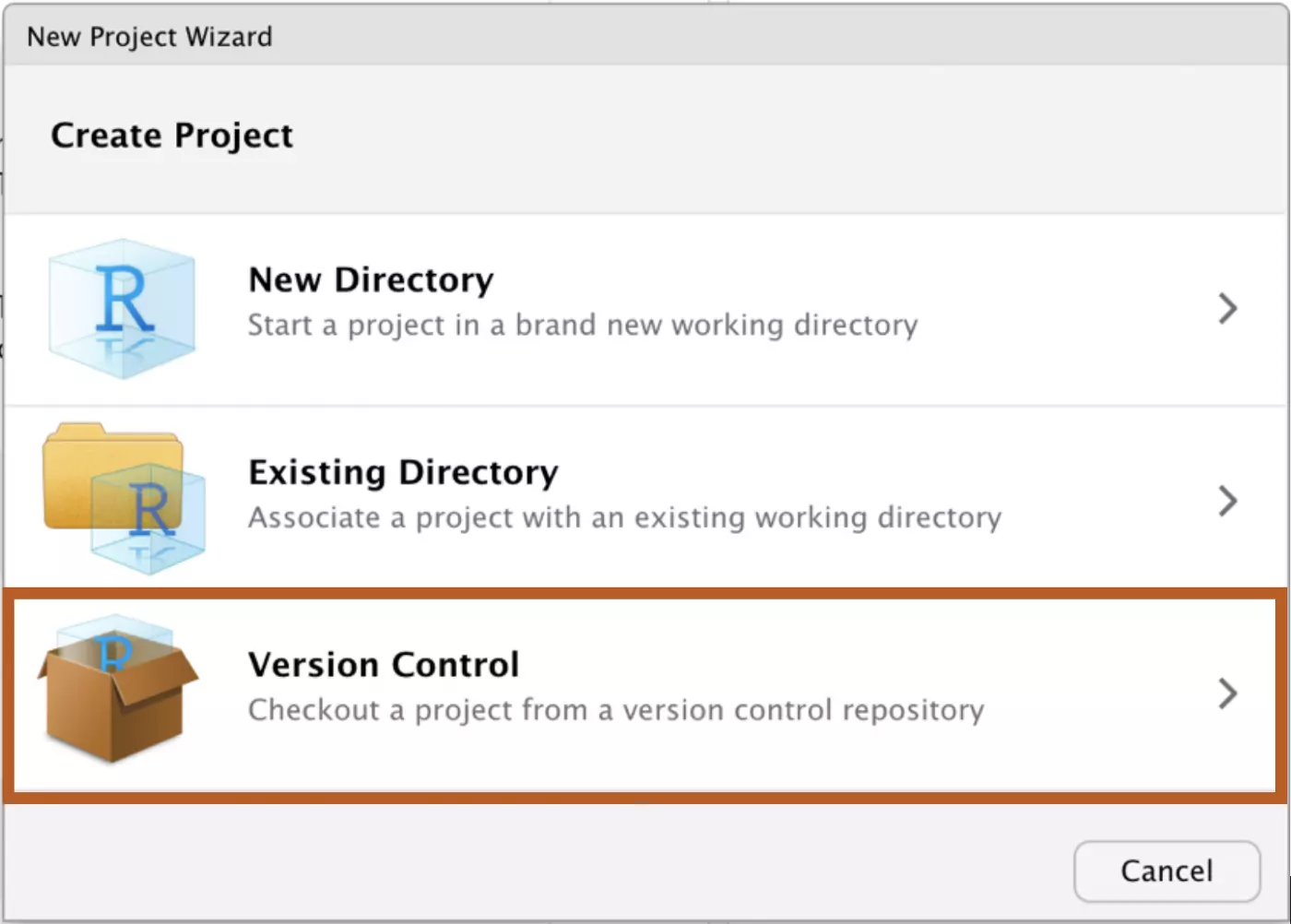
Select Git:
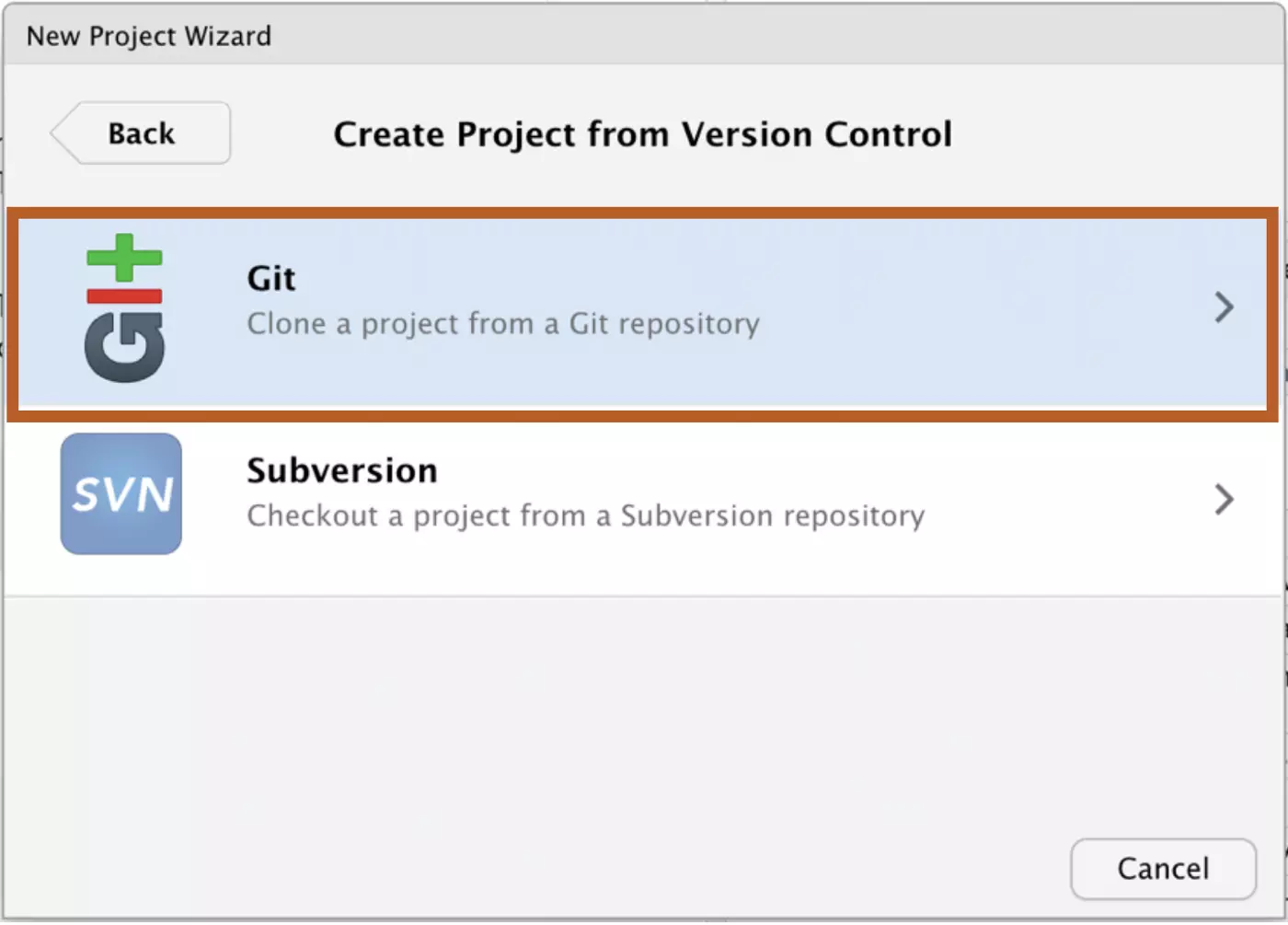
Finally, paste the URL that you copied from GitHub under Repository URL and click “Create Project”. I’ve created my project folder on my desktop, but you can browse to create your project anywhere on your computer.
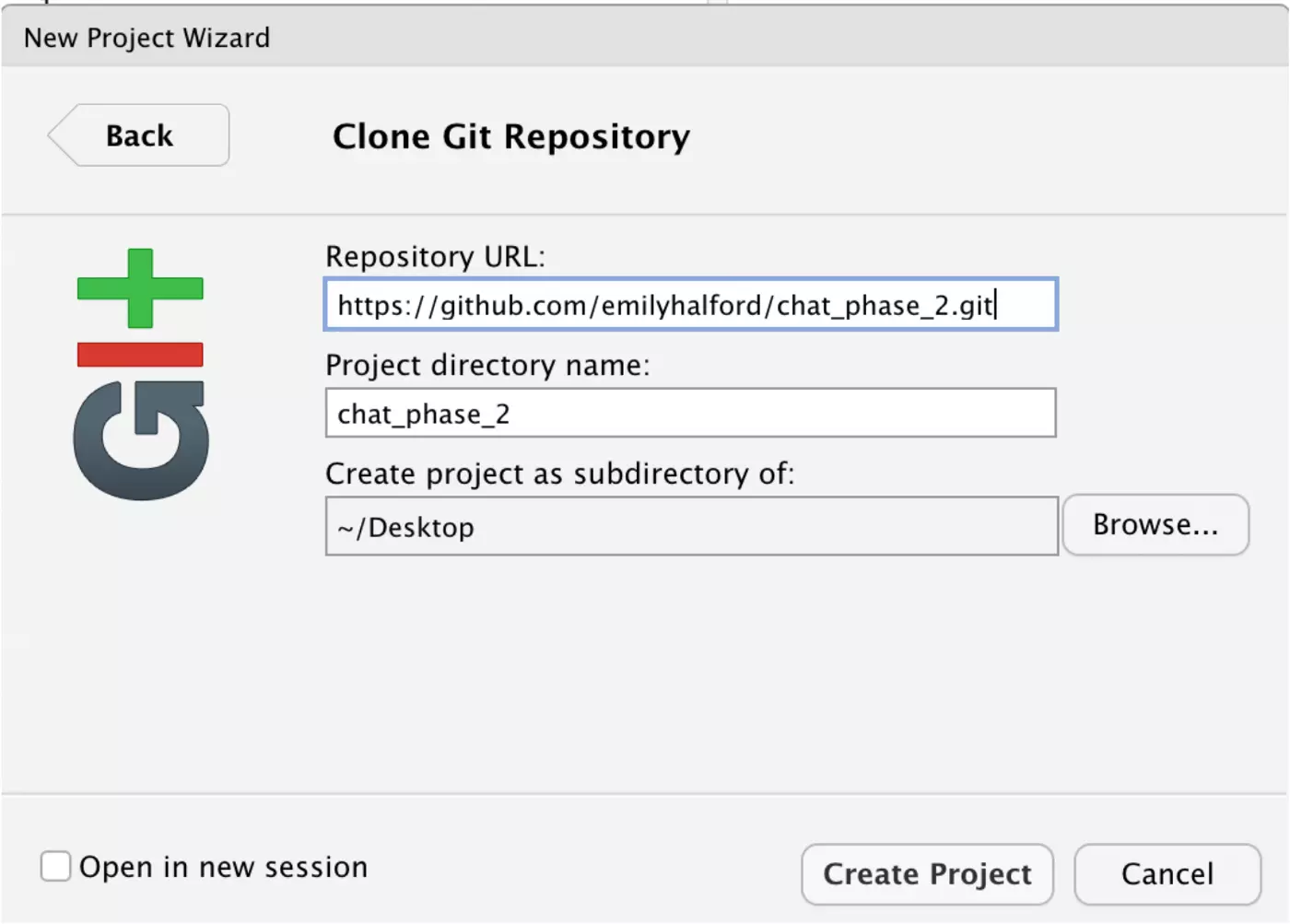
The project should open in R. You’ll know that it worked because your project will be indicated as the open project in the upper right corner of the screen, and you will now have a “Git” tab in the upper right panel.
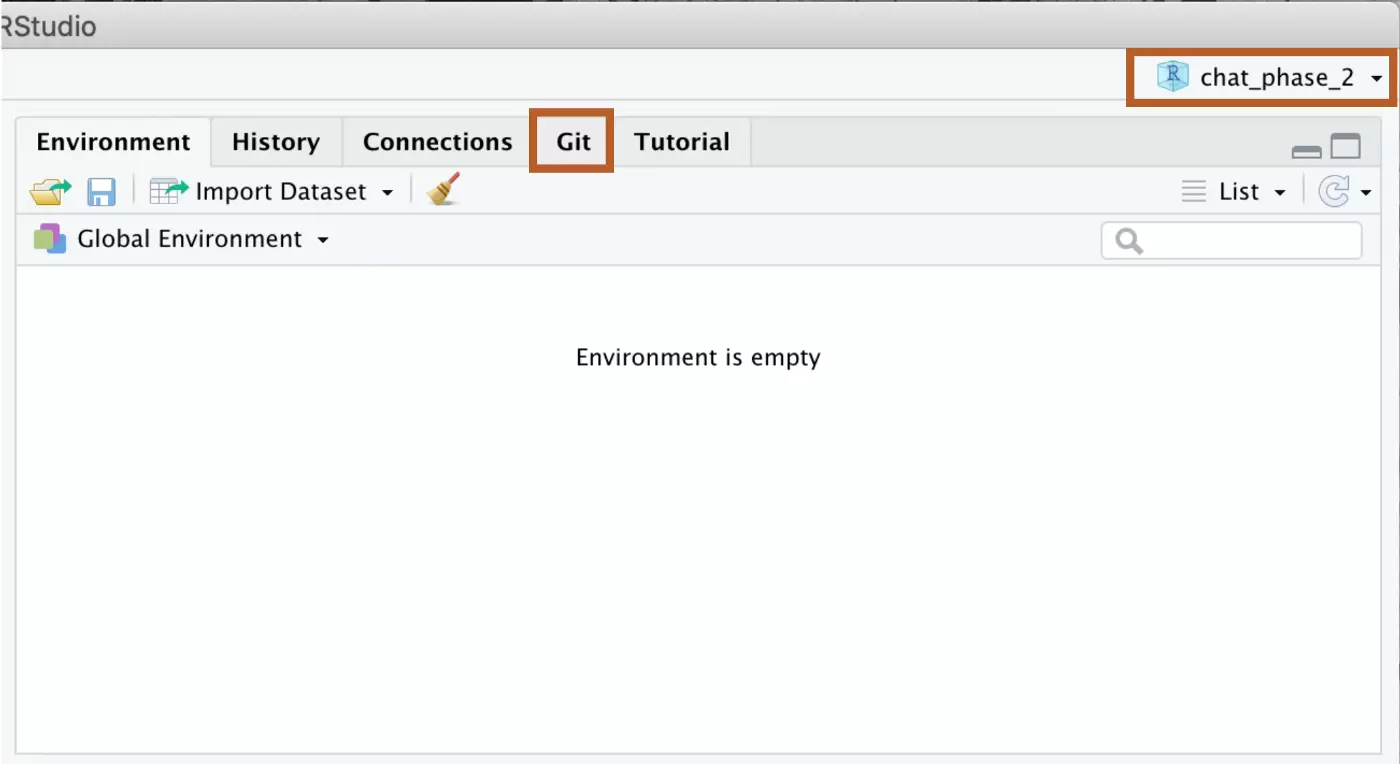
III. Store all related files in this project folder with meaningfully-named, consistent subfolders (such as a “data” folder that stores all of your data)
Now that you’ve created a local project and a GitHub repo, you’ll want to keep these clean and organized as you progress.
I added my data to the project in a data folder, so my current project folder looks like this:

As I progress, any additional data will be added to this “data” folder with consistent file names, I will create an “output” folder for any important graphs or other output that I create, and I will have several R Markdown files that are intentionally divided. For example, I may have one Markdown file where I clean data, one where I perform analyses, and one where I generate visualizations.
IV. Use the Git tab in R to consistently knit, commit, and push
GitHub only works for version control if you consistently push your work to their platform. In order to do this, click on the “Git” tab in RStudio. Any time you make changes to your local project, they’ll appear under this tab. First, click all of the changes that you’ve made since you last updated the repo. Next, click the “Commit” button.
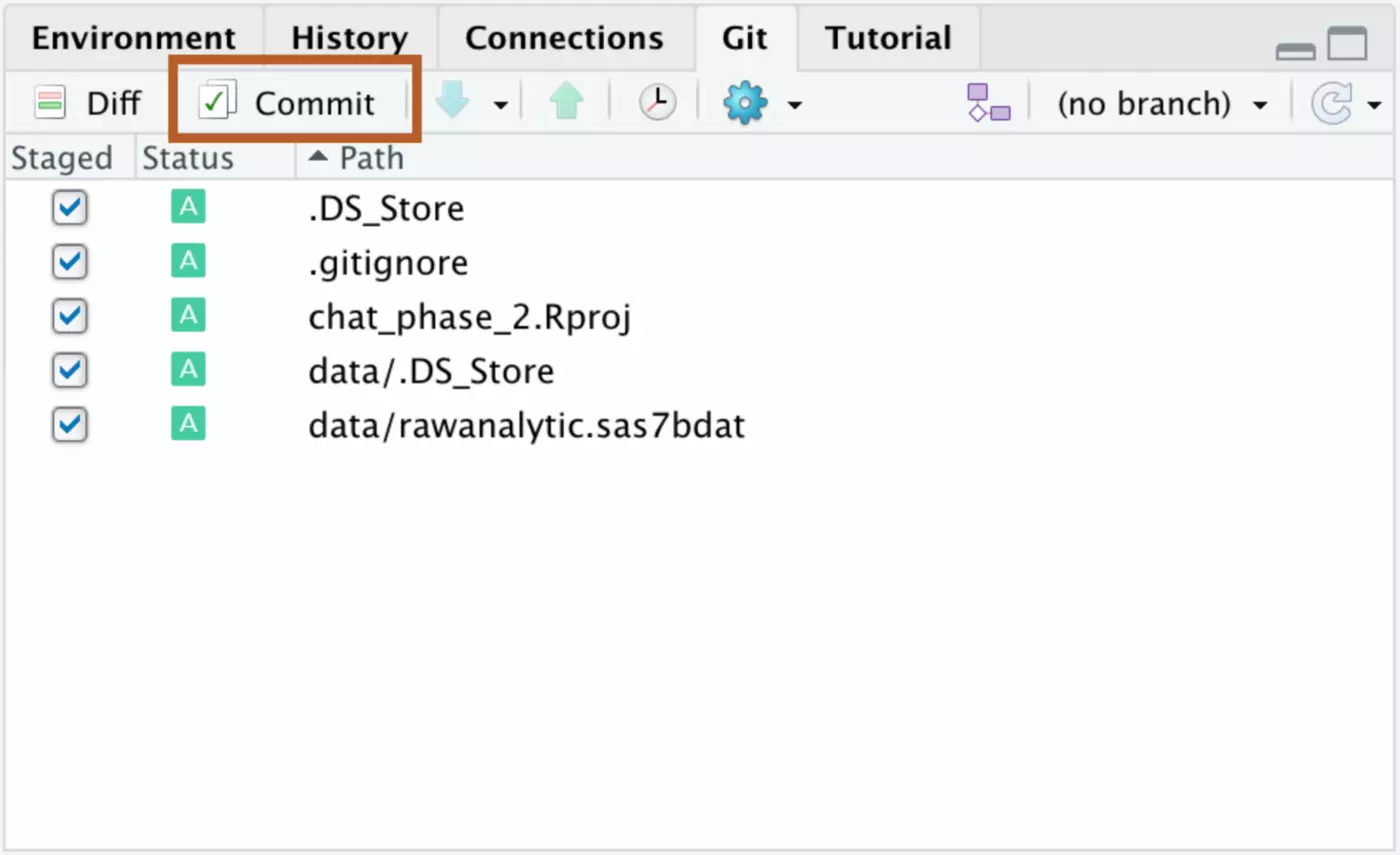
You will want to enter a commit message that describes the changes you are making. This makes it far easier for somebody else to navigate your repo, and will also make it easier to locate a previous version of your code if you need to access it later.
Once you’ve entered a commit message, select “Commit” in the bottom right corner. If that runs without error, the “Push” button will be accessible. Click “Push” to send your updates to GitHub.
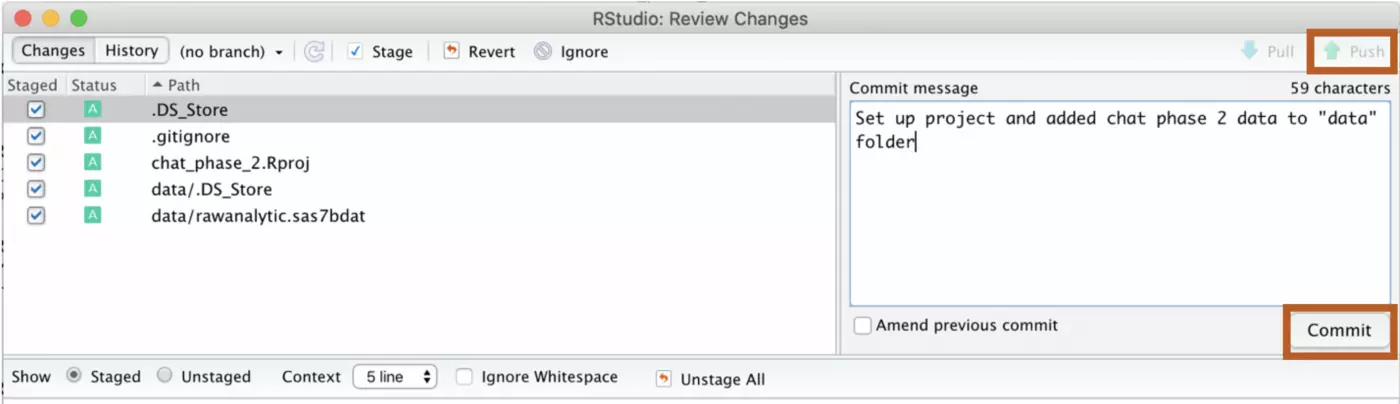
Now all of your changes are backed up on GitHub! You will want to do this consistently as you work. I recommend checking out your GitHub account the first time you do this to see how your work is stored there. If you are accessing somebody else’s repo, or if somebody else has made changes to yours, you can use “Pull” to pull the latest version of the repo to your computer.
5. Loading Packages
Now that everything is set up, you can start working in R!
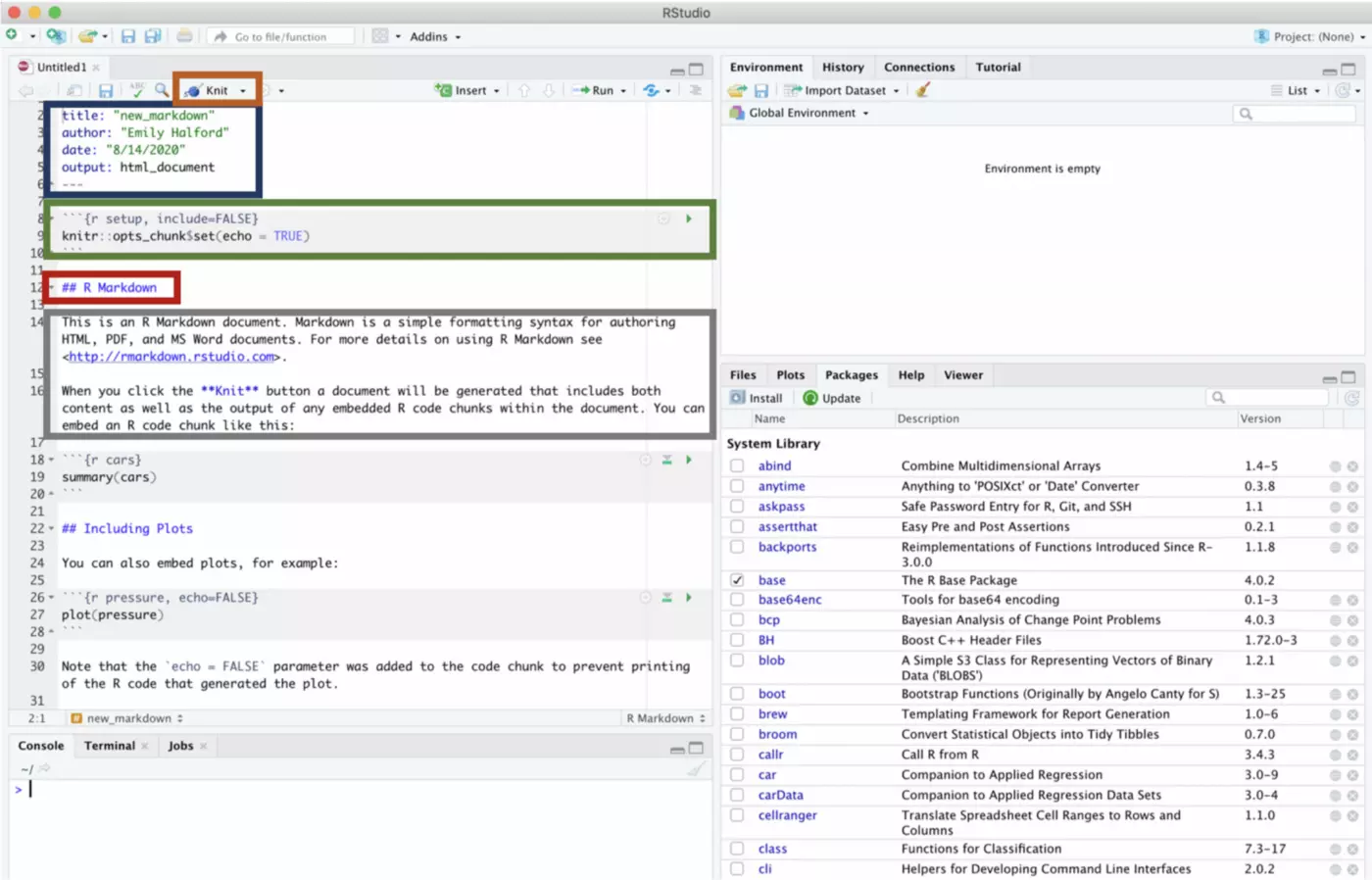
First, open up a new R Markdown file. If you click File → New File → R Markdown while your project is open (again, you can tell that your project is open if its name is in the top right of your window), the R Markdown file will be created in your project automatically.
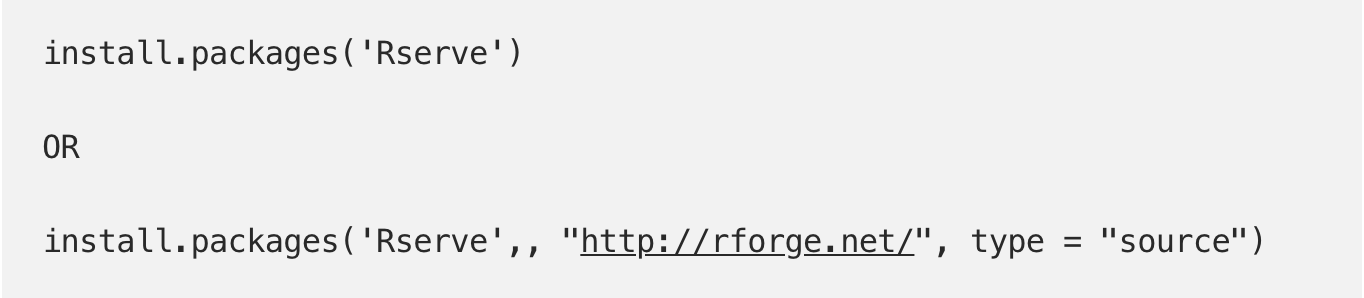
R automatically comes with “base R” functions. However, R is open-source (meaning that anyone can contribute packages), and there are a lot of amazing packages out there that you will need to use R efficiently. You will have to install these packages one time, and then load them every time that you want to use them. The code for doing so is provided below (note the double quotation marks to install a package and single quotes to load it). I like to install packages in the console, as I only have to do this once and therefore have no reason to save the line of code which installs the package. You will see confirmation in the console that the package has installed and loaded correctly.
Note: Tidyverse is actually a collection of packages rather than a single package. I highly, HIGHLY recommend familiarizing yourself with tidyverse and the packages included within it. They are widely used and are popular because they will make your life infinitely easier — I automatically load it every time I create a new Markdown file. You can find more information about tidyverse and its component packages here: https://www.tidyverse.org/
6. Other Notes
R is case-sensitive, unlike SAS and some other languages. This case-sensitivity is one of the reasons it’s so important to use consistent file- and variable-naming structures — you’ll be less likely to have annoying errors in your code and anyone else looking at your code will be more able to follow it. Personally, I use snake case (everything_is_lowercase_with_underscores) exclusively.
There are also established guidelines for how you should format your code. You should strive for code that not only runs, but is also tidy! One useful style guide from Hadley Wickham can be found here: http://adv-r.had.co.nz/Style.html
7. Up and Running!
You should now be able to open a new R Markdown file as part of an intentional workflow and to install and load packages within that file. These skills set you up to make even better use of the fantastic tutorials out there than can walk you through importing, cleaning, analyzing, and visualizing your data in R.


















Leave your comments
Post comment as a guest